
It is usually difficult to measure the relative importance of selection and neutral processes, including drift. The comparative importance of adaptive and non-adaptive forces in driving evolutionary change is an area of current research.
The neutral theory of molecular evolution proposed that most evolutionary changes are the result of the fixation of neutral mutations by genetic drift. Hence, in this model, most genetic changes in a population are the result of constant mutation pressure and genetic drift. This form of the neutral theory is now largely abandoned, since it does not seem to fit the genetic variation seen in nature. However, a more recent and better-supported version of this model is the nearly neutral theory, where a mutation that would be neutral in a small population is not necessarily neutral in a large population. Other alternative theories propose that genetic drift is dwarfed by other stochastic forces in evolution, such as genetic hitchhiking, also known as genetic draft.
The time for a neutral allele to become fixed by genetic drift depends on population size, with fixation occurring more rapidly in smaller populations. The number of individuals in a population is not critical, but instead a measure known as the effective population size. The effective population is usually smaller than the total population since it takes into account factors such as the level of inbreeding and the stage of the lifecycle in which the population is the smallest. The effective population size may not be the same for every gene in the same population.
Genetic Hitchhiking
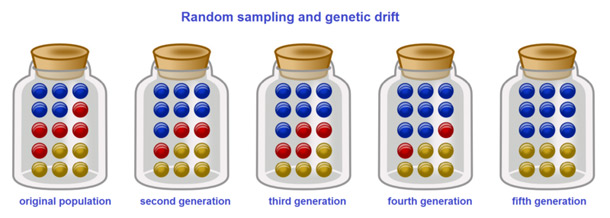
Analogies With Marble in a Jar
Repeat this process a number of times, randomly reproducing each generation of marbles to form the next. The numbers of red and blue marbles picked each generation fluctuates: sometimes more red, sometimes more blue. This fluctuation is genetic drift – a change in the population’s allele frequency resulting from a random variation in the distribution of alleles from one generation to the next.
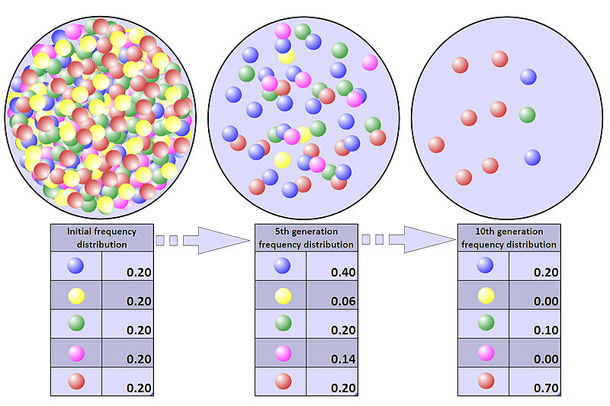
It is even possible that in any one generation no marbles of a particular color are chosen, meaning they have no offspring. In this example, if no red marbles are selected the jar representing the new generation contains only blue offspring. If this happens, the red allele has been lost permanently in the population, while the remaining blue allele has become fixed: all future generations are entirely blue. In small populations, fixation can occur in just a few generations.
Bottleneck Population
The impact of a population bottleneck can be sustained, even when the bottleneck is caused by a one-time event such as a natural catastrophe. After a bottleneck, inbreeding increases. This increases the damage done by recessive deleterious mutations, in a process known as inbreeding depression. The worst of these mutations are selected against, leading to the loss of other alleles that are genetically linked to them, in a process of background selection. This leads to a further loss of genetic diversity. In addition, a sustained reduction in population size increases the likelihood of further allele fluctuations from drift in generations to come.
A population’s genetic variation can be greatly reduced by a bottleneck, and even beneficial adaptations may be permanently eliminated. The loss of variation leaves the surviving population vulnerable to any new selection pressures such as disease, climate change or shift in the available food source, because adapting in response to environmental changes requires sufficient genetic variation in the population for natural selection to take place.
There have been many known cases of population bottleneck in the recent past. Prior to the arrival of Europeans, North American prairies were habitat for millions of greater prairie chickens. In Illinois alone, their numbers plummeted from about 100 million birds in 1900 to about 50 birds in the 1990s. The declines in population resulted from hunting and habitat destruction, but the random consequence has been a loss of most of the species’ genetic diversity. DNA analysis comparing birds from the mid century to birds in the 1990s documents a steep decline in the genetic variation in just in the latter few decades. Currently the greater prairie chicken is experiencing low reproductive success.
Over-hunting also caused a severe population bottleneck in the northern elephant seal in the 19th century. Their resulting decline in genetic variation can be deduced by comparing it to that of the southern elephant seal, which were not so aggressively hunted
Masel J (2011). “Genetic drift”. Current Biology 21 (20): R837–R838. doi:10.1016/j.cub.2011.08.007. PMID 22032182.
Futuyma, Douglas (1998). Evolutionary Biology. Sinauer Associates. p. Glossary. ISBN 0-87893-189-9.
Futuyma, Douglas (1998). Evolutionary Biology. Sinauer Associates. p. 320. ISBN 0-87893-189-9.
“Evolution 101:Sampling Error and Evolution”. University of California Berkeley. Retrieved 2009-11-01.
Walker J. “Introduction to Probability and Statistics”. The RetroPsychoKinesis Project. Fourmilab. Retrieved 2009-11-17.
Wahl L.M. (2011). “Fixation when N and s Vary: Classic Approaches Give Elegant New Results”. Genetics 188 (4): 783–785. doi:10.1534/genetics.111.131748. PMC 3176088. PMID 21828279.
Daniel Hartl, Andrew Clark (2007). Principles of Population Genetics, 4th edition. Sinauer Associates. p. 112. ISBN 978-0-87893-308-2.
Charlesworth B (March 2009). “Fundamental concepts in genetics: Effective population size and patterns of molecular evolution and variation”. Nat. Rev. Genet. 10 (3): 195–205. doi:10.1038/nrg2526. PMID 19204717.
Der R, Epstein CL, Plotkin JB (2011). “Generalized population models and the nature of genetic drift”. Theoretical Population Biology 80 (2): 80–99. doi:10.1016/j.tpb.2011.06.004. PMID 21718713.
Li, Wen-Hsiung; Dan Graur (1991). Fundamentals of Molecular Evolution. Sinauer Associates. p. 28. ISBN 0-87893-452-9.
Gillespie, John H. (2001). “Is the population size of a species relevant to its evolution?”. Evolution 55 (11): 2161–2169. PMID 11794777.
R.A. Neher and B.I. Shraiman (2011). “Genetic Draft and Quasi-Neutrality in Large Facultatively Sexual Populations”. Genetics 188 (4): 975–996. doi:10.1534/genetics.111.128876. PMC 3176096. PMID 21625002.
Warren Ewens (2004). Mathematical Population Genetics I. Theoretical Introduction. Interdisciplinary Applied Mathematics. Springer-Verlag.
Li, Wen-Hsiung; Dan Graur (1991). Fundamentals of Molecular Evolution. Sinauer Associates. p. 29. ISBN 0-87893-452-9.
Nicholas H. Barton, Derek E. G. Briggs, Jonathan A. Eisen, David B. Goldstein, Nipam H. Patel (2007). Evolution. Cold Spring Harbor Laboratory Press. p. 417. ISBN 0-87969-684-2.
Futuyma, Douglas (1998). Evolutionary Biology. Sinauer Associates. p. 300. ISBN 0-87893-189-9.
Otto S, Whitlock M (1 June 1997). “The Probability of Fixation in Populations of Changing Size”. Genetics 146 (2): 723–33. PMC 1208011. PMID 9178020.
Asher D. Cutter and Jae Young Choi (2010). “Natural selection shapes nucleotide polymorphism across the genome of the nematode Caenorhabditis briggsae”. Genome Research 20 (8): 1103–1111. doi:10.1101/gr.104331.109. PMC 2909573. PMID 20508143.
Hedrick, Philip W. (2004). Genetics of Populations. Jones and Bartlett Publishers. pp. 737. ISBN 0-7637-4772-6.
Wen-Hsiung Li, Dan Graur (1991). Fundamentals of Molecular Evolution. Sinauer Associates. p. 33. ISBN [[Special:BookSources/978-0-87893-452-9|978-0-87893-452-9]].
Kimura, Motoo; Ohta, Tomoko (2001). Theoretical Aspects of Population Genetics. Princeton University Press. pp. 232. ISBN
