Feline endogenous retroviruses
Cats (Felidae) present another example of virogene sequences in common descent. The standard phylogenetic tree for Felidae have smaller cats (Felis chaus, Felis silvestris, Felis nigripes, and Felis catus) diverging from larger cats such as the subfamily Pantherinae and other carnivores. The fact that small cats have an ERV where the larger cats do not suggests that the gene was inserted into the ancestor of the small cats after the larger cats had diverged.
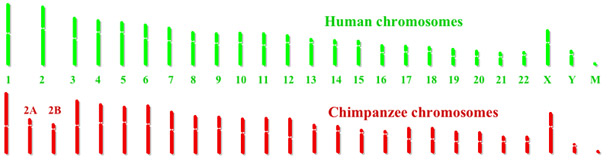
Chromosome 2 in humans
Fusion of ancestral chromosomes left distinctive remnants of telomeres, and a vestigial centromere
Evidence for the evolution of Homo sapiens from a common ancestor with chimpanzees is found in the number of chromosomes in humans as compared to all other members of Hominidae. All Hominidae (with the exception of humans) have 24 pairs of chromosomes. Humans have only 23 pairs. Human chromosome 2 is a result of an end-to-end fusion of two ancestral chromosomes.
The evidence for this includes:
- The correspondence of chromosome 2 to two ape chromosomes. The closest human relative, the common chimpanzee, has near-identical DNA sequences to human chromosome 2, but they are found in two separate chromosomes. The same is true of the more distant gorilla and orangutan.
- The presence of a vestigial centromere. Normally a chromosome has just one centromere, but in chromosome 2 there are remnants of a second centromere.
- The presence of vestigial telomeres. These are normally found only at the ends of a chromosome, but in chromosome 2 there are additional telomere sequences in the middle.
Chromosome 2 thus presents very strong evidence in favour of the common descent of humans and other apes. According to J. W. IJdo, “We conclude that the locus cloned in cosmids c8.1 and c29B is the relic of an ancient telomere-telomere fusion and marks the point at which two ancestral ape chromosomes fused to give rise to human chromosome 2.”
Genes of the Chromosome 2 fusion site
The results of the chimpanzee genome project suggest that when ancestral chromosomes 2A and 2B fused to produce human chromosome 2, no genes were lost from the fused ends of 2A and 2B. At the site of fusion, there are approximately 150,000 base pairs of sequence not found in chimpanzee chromosomes 2A and 2B. Additional linked copies of the PGML/FOXD/CBWD genes exist elsewhere in the human genome, particularly near the p end of chromosome 9. This suggests that a copy of these genes may have been added to the end of the ancestral 2A or 2B prior to the fusion event. It remains to be determined if these inserted genes confer a selective advantage.
- PGML. The phosphoglucomutase-like gene of human chromosome 2. This gene is incomplete and may not produce a functional transcript.
- FOXD. The forkhead box D4-like gene is an example of an intronless gene. The function of this gene is not known, but it may code for a transcription control protein.
- CBWD. Cobalamin synthetase is a bacterial enzyme that makes vitamin B12. In the distant past, a common ancestor to mice and apes incorporated a copy of a cobalamin synthetase gene (see: Horizontal gene transfer). Humans are unusual in that they have several copies of cobalamin synthetase-like genes, including the one on chromosome 2. It remains to be determined what the function of these human cobalamin synthetase-like genes is. If these genes are involved in vitamin B12 metabolism, this could be relevant to human evolution. A major change in human development is greater post-natal brain growth than is observed in other apes. Vitamin B12 is important for brain development, and vitamin B12 deficiency during brain development results in severe neurological defects in human children.
- CXYorf1-like protein. Several transcripts of unknown function corresponding to this region have been isolated. This region is also present in the closely related chromosome 9p terminal region that contains copies of the PGML/FOXD/CBWD genes.
Many ribosomal protein L23a pseudogenes are scattered through the human genome.
Cytochrome c
A classic example of biochemical evidence for evolution is the variance of the ubiquitous (i.e. all living organisms have it, because it performs very basic life functions) protein Cytochrome c in living cells. The variance of cytochrome c of different organisms is measured in the number of differing amino acids, each differing amino acid being a result of a base pair substitution, a mutation. If each differing amino acid is assumed the result of one base pair substitution, it can be calculated how long ago the two species diverged by multiplying the number of base pair substitutions by the estimated time it takes for a substituted base pair of the cytochrome c gene to be successfully passed on. For example, if the average time it takes for a base pair of the cytochrome c gene to mutate is N years, the number of amino acids making up the cytochrome c protein in monkeys differ by one from that of humans, this leads to the conclusion that the two species diverged N years ago.
The primary structure of cytochrome c consists of a chain of about 100 amino acids. Many higher order organisms possess a chain of 104 amino acids.
The cytochrome c molecule has been extensively studied for the glimpse it gives into evolutionary biology. Both chicken and turkeys have identical sequence homology (amino acid for amino acid), as do pigs, cows and sheep. Both humans and chimpanzees share the identical molecule, while rhesus monkeys share all but one of the amino acids: the 66th amino acid is isoleucine in the former and threonine in the latter.
What makes these homologous similarities particularly suggestive of common ancestry in the case of cytochrome C, in addition to the fact that the phylogenies derived from them match other phylogenies very well, is the high degree of functional redundancy of the cytochrome C molecule. The different existing configurations of amino acids do not significantly affect the functionality of the protein, which indicates that the base pair substitutions are not part of a directed design, but the result of random mutations that aren’t subject to selection.
Recent African origin of modern humans
Mathematical models of evolution, pioneered by the likes of Sewall Wright, Ronald Fisher and J. B. S. Haldane and extended via diffusion theory by Motoo Kimura, allow predictions about the genetic structure of evolving populations. Direct examination of the genetic structure of modern populations via DNA sequencing has allowed verification of many of these predictions. For example, the Out of Africa theory of human origins, which states that modern humans developed in Africa and a small sub-population migrated out (undergoing a population bottleneck), implies that modern populations should show the signatures of this migration pattern. Specifically, post-bottleneck populations (Europeans and Asians) should show lower overall genetic diversity and a more uniform distribution of allele frequencies compared to the African population. Both of these predictions are borne out by actual data from a number of studies.
Sources
Van Der Kuyl, AC; Dekker, JT; Goudsmit, J (1999). “Discovery of a New Endogenous Type C Retrovirus (FcEV) in Cats: Evidence for RD-114 Being an FcEVGag-Pol/Baboon Endogenous Virus BaEVEnv Recombinant”. Journal of Virology 73 (10): 7994–8002. PMC 112814. PMID 10482547.
Human Chromosome 2 is a fusion of two ancestral chromosomes by Alec MacAndrew; accessed 18 May 2006.
Evidence of Common Ancestry: Human Chromosome 2 (video) 2007
Yunis and Prakash; Prakash, O (1982). “The origin of man: a chromosomal pictorial legacy”. Science 215 (4539): 1525–1530. Bibcode 1982Sci…215.1525Y. doi:10.1126/science.7063861. PMID 7063861.
Human and Ape Chromosomes; accessed 8 September 2007.
Avarello, Rosamaria; Pedicini, A; Caiulo, A; Zuffardi, O; Fraccaro, M (1992). “Evidence for an ancestral alphoid domain on the long arm of human chromosome 2”. Human Genetics 89 (2): 247–9. doi:10.1007/BF00217134. PMID 1587535.
Ijdo, J. W.; Baldini, A; Ward, DC; Reeders, ST; Wells, RA (1991). “Origin of human chromosome 2: an ancestral telomere-telomere fusion”. Proceedings of the National Academy of Sciences 88 (20): 9051–5. Bibcode 1991PNAS…88.9051I. doi:10.1073/pnas.88.20.9051. PMC 52649. PMID 1924367.
Amino acid sequences in cytochrome c proteins from different species, adapted from Strahler, Arthur; Science and Earth History, 1997. page 348.
Lurquin PF, Stone L (2006). Genes, Culture, and Human Evolution: A Synthesis. Blackwell Publishing, Incorporated. p. 79. ISBN 1-4051-5089-0.
29+ Evidences for Macroevolution; Protein functional redundancy, Douglas Theobald, Ph.D.
Belshaw, R ; Pereira V; Katzourakis A; Talbot G; Paces J; Burt A; Tristem M. (2004). “Long-term reinfection of the human genome by endogenous retroviruses”. Proc Natl Acad Sci USA 101 (14): 4894–99. Bibcode 2004PNAS..101.4894B. doi:10.1073/pnas.0307800101. PMC 387345. PMID 15044706.
Bonner TI et al. (1982). “Cloned endogenous retroviral sequences from human DNA”. Proceedings of the National Academy of Sciences 79 (15): 4709–13. Bibcode 1982PNAS…79.4709B. doi:10.1073/pnas.79.15.4709. PMC 346746. PMID 6181510.
Pallen, Mark (2009). Rough Guide to Evolution. Rough Guides. pp. 200–206. ISBN 978-1-85828-946-5.
