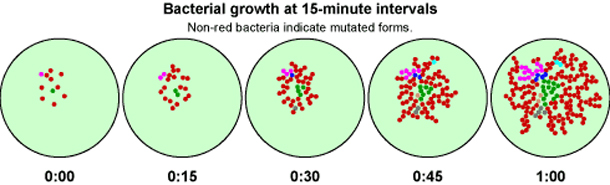
Antibiotic resistance can be a result of point mutations in the pathogen genome at a rate of about 1 in 108 per chromosomal replication. The antibiotic action against the pathogen can be seen as an environmental pressure; those bacteria which have a mutation allowing them to survive will live on to reproduce. They will then pass this trait to their offspring, which will result in a fully resistant colony.
Understanding the changes that have occurred during organism’s evolution can reveal the genes needed to construct parts of the body, genes which may be involved in human genetic disorders. For example, the Mexican tetra is an albino cavefish that lost its eyesight during evolution. Breeding together different populations of this blind fish produced some offspring with functional eyes, since different mutations had occurred in the isolated populations that had evolved in different caves. This helped identify genes required for vision and pigmentation, such as crystallins and the melanocortin 1 receptor. Similarly, comparing the genome of the Antarctic icefish, which lacks red blood cells, to close relatives such as the Antarctic rockcod revealed genes needed to make these blood cells.
The Escape of the Pathogens: An Evolutionary Arms Race
Human populations are constantly locked in evolutionary arms races with pathogens that invade our bodies. We must recognize that these pathogens (such as the flu virus shown at right) are continuously evolving entities in order to develop better ways to fight them and control their evolution.An ounce of prevention…every year?
Recently, the mayor of New York City called upon citizens to get a head start on one particular evolutionary arms race: “I urge older New Yorkers and others at risk to protect themselves from flu and pneumonia through a simple and proven ounce of prevention: immunizations. The time to get immunized is now, before the peak of the flu season.”
Many of those New Yorkers had already gotten flu shots the year before and the year before that, but, perhaps strangely, they were being asked to get yet another immunization. Why do we need a new flu shot every year? Can’t modern medicine invent just one vaccine that would do the trick?
Flu viruses evolve rapidly.
As they circulate through populations around the world and switch hosts, flu viruses change so much that our vaccines are rendered obsolete every year. The flu is a problem for which a solution must be redesigned and rebuilt every year, like a bridge that gets washed away every flood season. Only by understanding the flu as an evolving entity can we understand why our solution to the problem must change every year.
Every day we come into contact with millions of bacteria and viruses. Some are harmful and others are beneficial, while the rest have no apparent effect on our health. When harmful microorganisms enter our bodies, a battle ensues.
Rapid reproduction and natural selection
Because bacteria and viruses reproduce rapidly, they evolve rapidly. These short generation times — some bacteria have a generation time of just 15 minutes — mean that natural selection acts quickly. In each pathogen generation, new mutations and gene combinations are generated that then pass through the selective filter of our drugs and immune response. Over the course of many pathogen generations (a small fraction of a single human lifetime), they adapt to our defenses, evolving right out from under our attempts to rid ourselves of them.
Applying our knowledge of evolution
But that doesn’t mean that we should stop trying to win these battles. By understanding these pathogens as evolving entities, subject to the same processes of evolution that we can study in fruit flies or the fossil record, we may be able to identify ways to slow their progress.
courtesy of UC Berkeley
Antibiotic Resistance: Delaying the Inevitable
Only a few decades ago, antibiotics were considered to be wonder drugs because they worked so well to cure deadly diseases. Ironically, though, many antibiotics have become less effective, precisely because they have worked so well and have been used so often.Making inroads against infectious disease
The antibiotic era began in 1929 with Alexander Fleming’s observation that bacteria would not grow near colonies of the mold Penicillium. In the decades that followed this breakthrough discovery, molecules produced by fungi and bacteria have been successfully used to combat bacterial diseases such as tuberculosis and pneumonia. Antibiotics drastically reduced death rates associated with many infectious diseases.
Infectious diseases strike back
The golden age of antibiotics proved to be a short-lived one. During the past few decades, many strains of bacteria have evolved resistance to antibiotics. An example of this is Neisseria gonorrhoeae, the bacteria that causes gonorrhea, shown at right. In the 1960s penicillin and ampicillin were able to control most cases of gonorrhea. Today, more than 24 percent of gonorrheal bacteria in the U.S. are resistant to at least one antibiotic, and 98 percent of gonorrheal bacteria in Southeast Asia are resistant to penicillin. Infectious bacteria are much harder to control than their predecessors were ten or twenty years ago.
Doctors miss the “good old days,” when the antibiotics they prescribed consistently cured their patients. However, evolutionary theory suggests some specific tactics to help slow the rate at which bacteria become resistant to our drugs.
Applying our knowledge of evolution
Evolutionary theory predicted that bacterial resistance would happen. Given time, heredity, and variation, any living organisms (including bacteria) will evolve when a selective pressure (like an antibiotic) is introduced. But evolutionary theory also gives doctors and patients some specific strategies for delaying even more widespread evolution of antibiotic resistance. These strategies include:
1. Don’t use antibiotics to treat viral infections.
Antibiotics kill bacteria, not viruses. If you take antibiotics for a viral infection (like a cold or the flu), you will not kill the viruses, but you will introduce a selective pressure on bacteria in your body, inadvertently selecting for antibiotic-resistant bacteria. Basically, you want your bacteria to be “antibiotic virgins,” so that if they someday get out of hand and cause an infection that your immune system can’t handle, they can be killed by a readily available antibiotic.
2. Avoid mild doses of antibiotics over long time periods.
If an infection needs to be controlled with antibiotics, a short-term, high-dosage prescription is preferable. This is because you want to kill all of the illness-causing bacteria, leaving no bacterial survivors. Any bacteria that survive a mild dose are likely to be somewhat resistant. Basically, if you are going to introduce a selective pressure (antibiotics), make it so strong that you cause the extinction of the illness-causing bacteria in the host and not their evolution into resistant forms.
3. When treating a bacterial infection with antibiotics, take all your pills.
Just as mild doses can breed resistance, an incomplete regimen of antibiotics can let bacteria survive and adapt. If you are going to introduce a selective pressure (antibiotics), make it a really strong one and a long enough one to cause the extinction of the illness-causing bacteria and not their evolution.
4. Use a combination of drugs to treat a bacterial infection.
If one particular drug doesn’t help with a bacterial infection, you may be dealing with a resistant strain. Giving a stronger dose of the same antibiotic just increases the strength of the same selective pressure — and may even cause the evolution of a “super-resistant” strain. Instead, you might want to try an entirely different antibiotic that the bacteria have never encountered before. This new and different selective pressure might do a better job of causing their extinction, not their evolution.
5. Reduce or eliminate the “preventive” use of antibiotics on livestock and crops.
Unnecessary use of antibiotics for agricultural and livestock purposes may lead to the evolution of resistant strains. Later, these strains will not be able to be controlled by antibiotics when it really is necessary. Preventive use of antibiotics on livestock and crops can also introduce antibiotics into the bodies of the humans who eat them.
Ultimately, recognizing bacteria as evolving entities and understanding their evolution should help us to control that evolution, allowing us to prolong the useful lifespan of antibiotics.
courtesy of UC Berkeley
Huntington’s Chorea: Evolution and Genetic Disease
Huntington’s chorea is a devastating human genetic disease. A close look at its genetic origins and evolutionary history explains its persistence and points to a potential solution to this population-level problem.People who inherit this genetic disease have an abnormal dominant allele that disrupts the function of their nerve cells, slowly eroding their control over their bodies and minds and ultimately leading to death. In the fishing villages located near Lake Maracaibo in Venezuela, there are more people with Huntington’s disease than anywhere else in the world. In some villages, more than half the people may develop the disease.
How is it possible that such a devastating genetic disease is so common in some populations? Shouldn’t natural selection remove genetic defects from human populations? Research on the evolutionary genetics of this disease suggests that there are two main reasons for the persistence of Huntington’s in human populations: mutation coupled with weak selection.
Mutation
In 1993, a collaborative research group discovered the culprit responsible for Huntington’s: a stretch of DNA that repeats itself over and over again, CAGCAGCAGCAG… and so on. People carrying too many CAGs in the Huntington’s gene (more than about 35 repeats) develop the disease. In most cases, those affected by Huntington’s inherited a disease-causing allele from a parent. Others may have no family history of the disease, but may have new mutations which cause Huntington’s.
If a mutation ends up inserting extra CAGs into the Huntington’s gene, new Huntington’s alleles may be created. Of course it’s also possible for a mutation to remove CAGs. But research suggests that for Huntington’s, mutation is biased; additions of CAGs are more likely than losses of CAGs.
Selection
As though that weren’t bad enough, Huntington’s belongs to a class of genetic diseases that largely escape natural selection. Huntington’s is often “invisible” to natural selection for a very simple reason: it generally does not affect people until after they’ve reproduced. In this way, the alleles for late-onset Huntington’s may evade natural selection, “sneaking” into the next generation, despite its deleterious effects. Early-onset cases of Huntington’s are rare; these are an exception, and are strongly selected against.
Persistence
These mechanisms of evolution, mutation and selection, can help us understand the persistence of Huntington’s in populations. In general, Huntington’s is rare — 30-70 cases per million people in most Western countries — but it is not entirely eliminated because selection does a relatively poor job of weeding these alleles out, while mutation continues creating new ones.
Dr. Nancy Wexler has been studying the remarkably high frequency of Huntington’s in Lake Maracaibo since the 1970s. She has found that the high incidence of this disease there is explained by an evolutionary event called the founder effect. About 200 years ago, a single woman who happened to carry the Huntington’s allele bore 10 children — and today, many residents of Lake Maracaibo trace their ancestry (and their disease-causing gene) back to this lineage. A simple fluke of history, high-birth rates, and weak selection are responsible for the genetic burden shouldered by this population.
Solutions?
Currently, physicians don’t have any cures for Huntington’s disease — there’s no miracle pill that will stop the progress of the disease. However, understanding the evolutionary history of the disease — a recurrent mutation that is often “missed” by natural selection — points out a way to reduce the frequency of the disease in the long term: allowing people to make more informed reproductive choices.
Today, genetic testing can identify people who carry a Huntington’s allele long before the onset of the disease and before they have made their reproductive choices. The genetic test that identifies the Huntington’s allele works sort of like DNA fingerprinting. A DNA sample is copied and cut into pieces. The pieces are then spread out on a gel. The banding pattern can tell researchers whether a person carries an allele that is likely to cause Huntington’s.

Having this information could allow people to make more-informed reproductive decisions. For example, at Lake Maracaibo, researchers and health workers have tried to make contraception available to the local population so that they can make reproductive choices based on their own family history with the disease. But whatever people eventually decide to do with this knowledge, a deep understanding of the disease would not be possible without the historical perspective offered by evolution.
courtesy of UC Berkeley
Understanding Evolution is Important to Medicine
Understanding evolution helps us solve biological problems that impact our lives. There are excellent examples of this in the field of medicine. To stay one step ahead of pathogenic diseases, researchers must understand the evolutionary patterns of disease-causing organisms. To control hereditary diseases in people, researchers study the evolutionary histories of the disease-causing genes. In these ways, a knowledge of evolution can improve the quality of human life.courtesy of UC Berkeley
Evolution and Medicine: Helpful Links
Darwinian medicine: Applications of evolutionary biology for veterinariansRelevance of evolution: medicine
Sources
Maher B. (2009). “Evolution: Biology’s next top model?”. Nature 458 (7239): 695–8. doi:10.1038/458695a. PMID 19360058.
Borowsky R (2008). “Restoring sight in blind cavefish”. Curr. Biol. 18 (1): R23–4. doi:10.1016/j.cub.2007.11.023. PMID 18177707.
Gross JB, Borowsky R, Tabin CJ (2009). “A novel role for Mc1r in the parallel evolution of depigmentation in independent populations of the cavefish Astyanax mexicanus”. PLoS Genet. 5 (1): e1000326. doi:10.1371/journal.pgen.1000326. PMC 2603666. PMID 19119422.
Yergeau DA, Cornell CN, Parker SK, Zhou Y, Detrich HW (2005). “bloodthirsty, an RBCC/TRIM gene required for erythropoiesis in zebrafish”. Dev. Biol. 283 (1): 97–112. doi:10.1016/j.ydbio.2005.04.006. PMID 15890331
