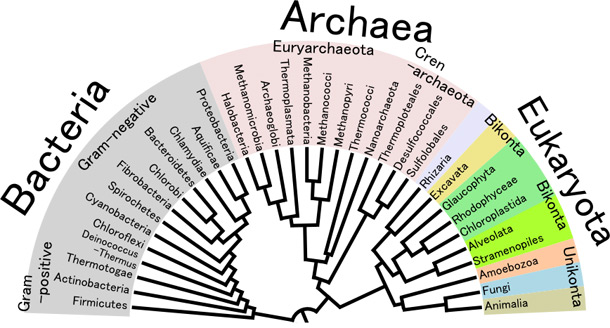
The simplest and most powerful evidence is provided by phylogenetic reconstruction. Such reconstructions, especially when done using slowly evolving protein sequences, are often quite robust and can be used to reconstruct a great deal of the evolutionary history of modern organisms (and even in some instances such as the recovered gene sequences of mammoths, Neanderthals or T. rex, the evolutionary history of extinct organisms). These reconstructed phylogenies recapitulate the relationships established through morphological and biochemical studies. The most detailed reconstructions have been performed on the basis of the mitochondrial genomes shared by all eukaryotic organisms, which are short and easy to sequence; the broadest reconstructions have been performed either using the sequences of a few very ancient proteins or by using ribosomal RNA sequence.
Phylogenetic relationships also extend to a wide variety of nonfunctional sequence elements, including repeats, transposons, pseudogenes, and mutations in protein-coding sequences that do not result in changes in amino-acid sequence. While a minority of these elements might later be found to harbor function, in aggregate they demonstrate that identity must be the product of common descent rather than common function.
DNA Sequencing and Phylogenetic Trees
Endogenous Retroviruses
Proteins
Mount DM. (2004). Bioinformatics: Sequence and Genome Analysis (2nd ed.). Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY.. ISBN 0-87969-608-7.
Douglas J. Futuyma (1998). Evolutionary Biology (3rd ed.). Sinauer Associates Inc.. pp. 108–110. ISBN 0-87893-189-9.
Haszprunar (1995). “The mollusca: Coelomate turbellarians or mesenchymate annelids?”. In Taylor. Origin and evolutionary radiation of the Mollusca : centenary symposium of the Malacological Society of London. Oxford: Oxford Univ. Press. ISBN 0-19-854980-6.
Kozmik, Z; Daube, M; Frei, E; Norman, B; Kos, L; Dishaw, LJ; Noll, M; Piatigorsky, J (2003). “Role of Pax genes in eye evolution: A cnidarian PaxB gene uniting Pax2 and Pax6 functions”. Developmental cell 5 (5): 773–85. doi:10.1016/S1534-5807(03)00325-3. PMID 14602077.
Kozmik, Z; Daube, Michael; Frei, Erich; Norman, Barbara; Kos, Lidia; Dishaw, Larry J.; Noll, Markus; Piatigorsky, Joram (2003). “Role of Pax Genes in Eye Evolution A Cnidarian PaxB Gene Uniting Pax2 and Pax6 Functions”. Developmental Cell 5 (5): 773–785. doi:10.1016/S1534-5807(03)00325-3. PMID 14602077.
Land, M.F. and Nilsson, D.-E., Animal Eyes, Oxford University Press, Oxford (2002) ISBN 0-19-850968-5.
Chen FC, Li WH (2001). “Genomic Divergences between Humans and Other Hominoids and the Effective Population Size of the Common Ancestor of Humans and Chimpanzees”. Am J Hum Genet. 68 (2): 444–56. doi:10.1086/318206. PMC 1235277. PMID 11170892.
Cooper GM, Brudno M, Green ED, Batzoglou S, Sidow A (2003). “Quantitative Estimates of Sequence Divergence for Comparative Analyses of Mammalian Genomes”. Genome Res. 13 (5): 813–20. doi:10.1101/gr.1064503. PMC 430923. PMID 12727901.
The picture labeled “Human Chromosome 2 and its analogs in the apes” in the article Comparison of the Human and Great Ape Chromosomes as Evidence for Common Ancestry is literally a picture of a link in humans that links two separate chromosomes in the nonhuman apes creating a single chromosome in humans. Also, while the term originally referred to fossil evidence, this too is a trace from the past corresponding to some living beings that, when alive, physically embodied this link.
The New York Times report Still Evolving, Human Genes Tell New Story, based on A Map of Recent Positive Selection in the Human Genome, states the International HapMap Project is “providing the strongest evidence yet that humans are still evolving” and details some of that evidence.
“29+ Evidences for Macroevolution: The Scientific Case for Common Descent”. Theobald, Douglas. Retrieved 2011-03-10.
“Converging Evidence for Evolution.” Phylointelligence: Evolution for Everyone. Web. 26 Nov. 2010.
Petrov DA, Hartl DL (2000). “Pseudogene evolution and natural selection for a compact genome”. J Hered. 91 (3): 221–7. doi:10.1093/jhered/91.3.221. PMID 10833048
